Fig. 1
From: XL-DNase-seq: improved footprinting of dynamic transcription factors
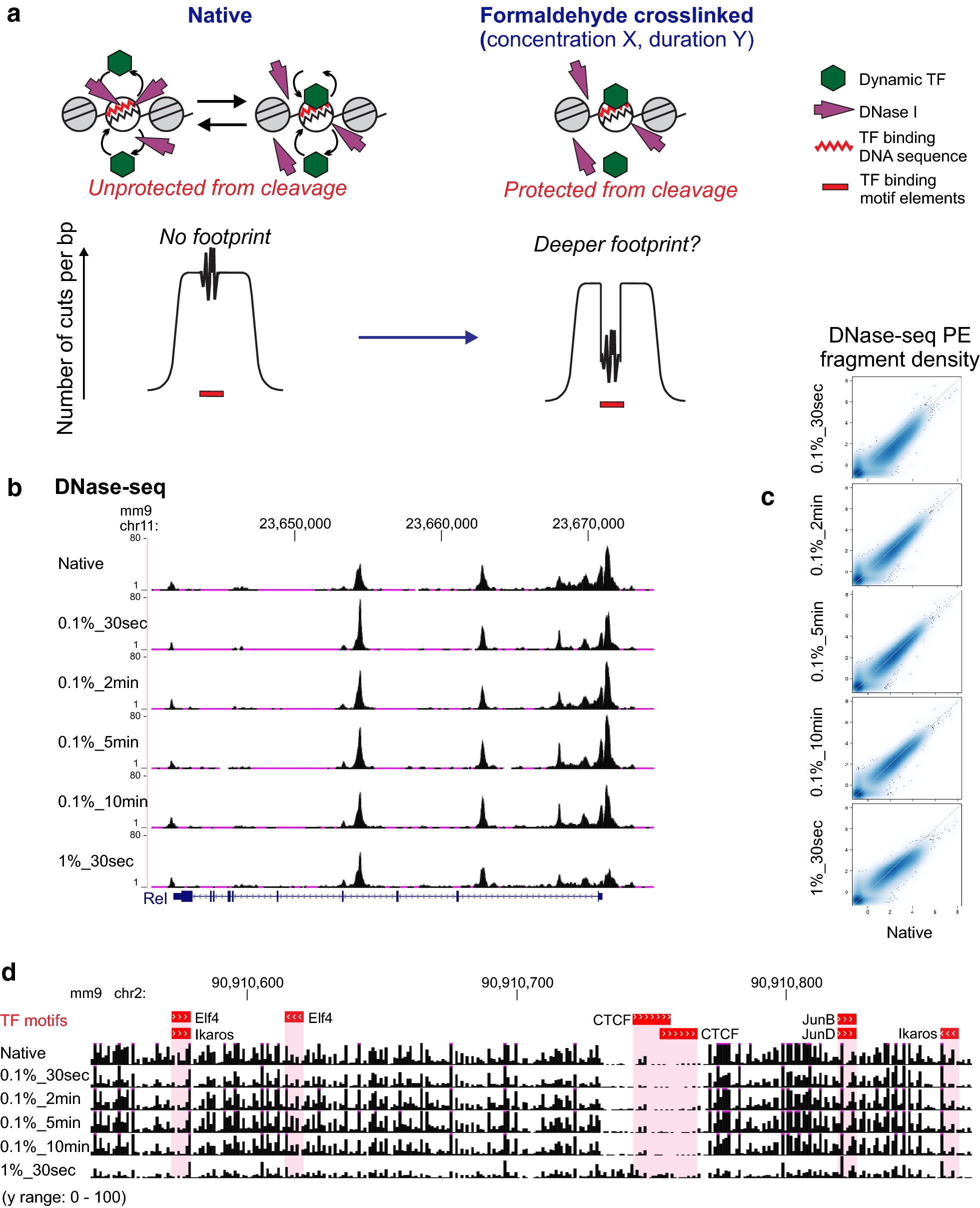
XL-DNase-seq is designed to capture short-lived binding of TFs and preserve chromatin accessibility profiles. a Rationale for introducing a cross-linking step prior to digestion by DNase I. b Near-identical DNase-seq density profiles across cross-linking conditions. XL-DNase-seq preserves the chromatin accessibility profile as observed by the native DNase-seq. The genome browser shot of the Rel locus shows DNase-seq signal tracks from all cross-linking conditions. c Fragment density normalized to 10 million mapped reads. Genome-wide comparison of DNase-seq fragment density profiles across the cross-linking conditions. d A browser shot of cross-linked raw cut count tracks. The top track shows the locations of TF binding motif elements obtained by FIMO. Reference genome: mm9. See also Additional file 2: Fig. S1
