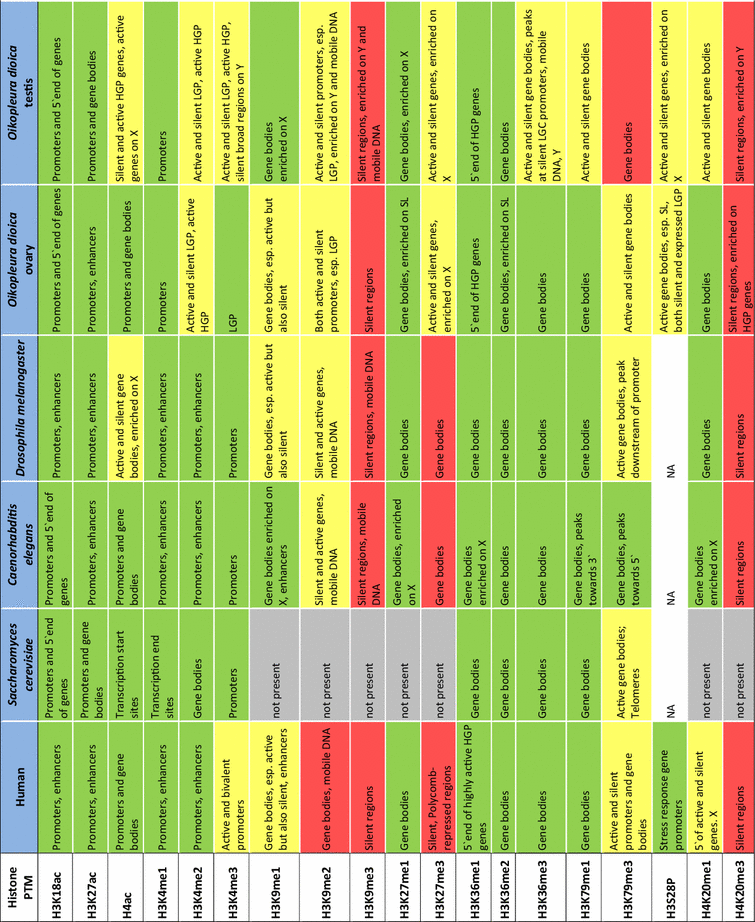
- Assignments of features marked by histone PTMs for species other than Oikopleura dioica were based on reference literature as follows: human [12, 13, 46–49]; Saccharomyces cerevisiae [50–52]; Caenorhabditis elegans [12, 14, 15]; Drosophila melanogaster [8, 10, 12]. Associations of individual histone modification Oikopleura dioica were assigned based on assessment of enrichment plots shown in Supplemental Figure S1. These plots show the mean signal intensity around gene start and end sites with genes grouped according to their expression level, the GC content of their promoter, their trans-splicing status and chromosomal locations. We used the error bars indicating 95% confidence intervals on the mean to define (by the lack of their overlap) relative enrichments between compared gene sets as significant. Colors indicate transcriptional activity of features marked by PTMs: green = transcribed; red = repressed; yellow = both active and silent. Esp. = especially; X = X-chromosome; Y = Y-chromosome; HGP/LGP = high/low GC content promoter; SL genes = genes whose transcripts are subject to trans-splicing of the splice leader (SL) sequence; N = not analyzed

