Fig. 2
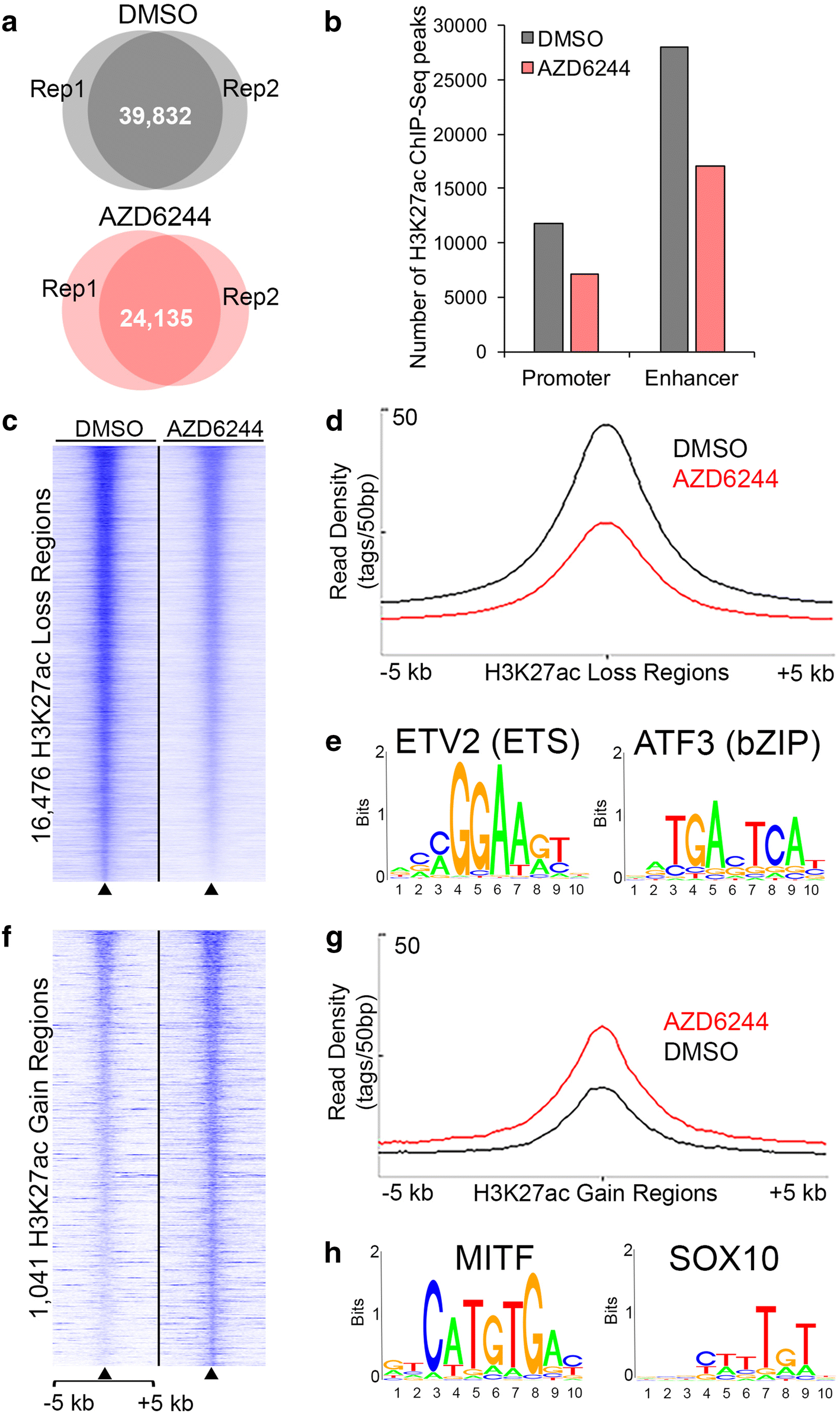
MEK inhibition induces genome-wide modulation of the active chromatin landscape in 501mel cells. a Venn diagrams showing overlap of H3K27ac peaks identified by ChIP-Seq in independent replicate samples of 501mel cells treated with DMSO (upper panel) and AZD6244 (lower panel). b Distribution of H3K27ac peaks in 501mel cells treated with DMSO (gray) or AZD6244 (red) at either promoter (< 2.5 kb from TSS) or non-promoter, distal enhancer regions. c, f Heatmaps showing relative H3K27ac signal in 501mel cells treated with DMSO or AZD6244. H3K27ac ChIP-Seq tag densities are shown for significantly differentially remodeled H3K27ac sites (FDR < 0.05) at regions c losing H3K27ac or f gaining H3K27ac. Each row represents a 10-kb region centered on differentially remodeled H3K27ac regions, sorted by H3K27ac signal enrichment. d, g Line plots show average H3K27ac ChIP-Seq signal intensities (y-axis) across regions d losing H3K27ac or g gaining H3K27ac over a ± 5 kb genomic window centered on differentially remodeled H3K27ac sites (x-axis). e, h The top significantly enriched transcription factor motifs identified by de novo motif discovery of sequences in regions e losing H3K27ac and h gaining H3K27ac. The top 5 enriched motifs, the fraction of H3K27ac regions containing the motifs, and the p values for the overrepresentation of each motif are shown in Additional file 2: Table S3
